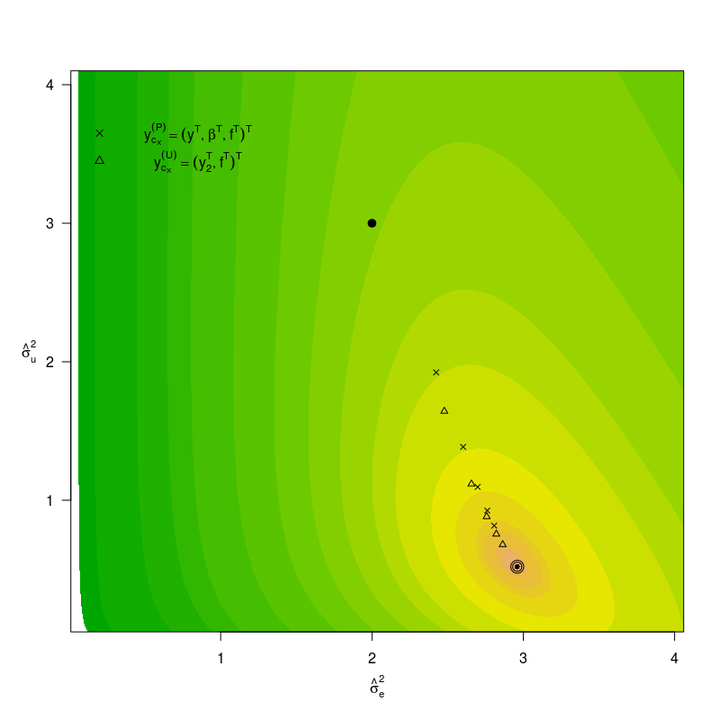
Abstract
Linear mixed models are regularly applied to animal and plant breeding data to evaluate genetic potential. Residual maximum likelihood (REML) is the preferred method for estimating variance parameters associated with this type of model. Typically an iterative algorithm is required for the estimation of variance parameters. Two algorithms which can be used for this purpose are the expectation‐maximisation (EM) algorithm and the parameter expanded EM (PX‐EM) algorithm. Both, particularly the EM algorithm, can be slow to converge when compared to a Newton‐Raphson type scheme such as the average information (AI) algorithm. The EM and PX‐EM algorithms require specification of the complete data, including the incomplete and missing data. We consider a new incomplete data specification based on a conditional derivation of REML. We illustrate the use of the resulting new algorithm through two examples; a sire model for lamb weight data and a balanced incomplete block soybean variety trial. In the cases where the AI algorithm failed, a REML PX‐EM based on the new incomplete data specification converged in 28% to 30% fewer iterations than the alternative REML PX‐EM specification. For the soybean example a REML EM algorithm using the new specification converged in fewer iterations than the current standard specification of a REML PX‐EM algorithm. The new specification integrates linear mixed models, Henderson’s mixed model equations, REML and the REML EM algorithm into a cohesive framework.